VirusCommander#
Tool for entering new viruses, batches of viruses, opsins, promotors etc. into the Database. Having the viruses in the database and associated with animals via surgey will allow easy search what was done with any given virus and with what results.
Entering a virus or virus batch here will automatically push it also to eLabFTW.
From the datastructure_tools directory run
python ./VirusCommander.py
or from any directory
python -m datastructure_tools virus
Error
In case you get this error, no connection can be established to MySQL. Ensure you entered correct credentials first using AdminCommander.
Add a new virus to DB#
A new virus can be added and connected to its properties such as promots, opsin etc. Option you need is not available? Add new one as indicated later.
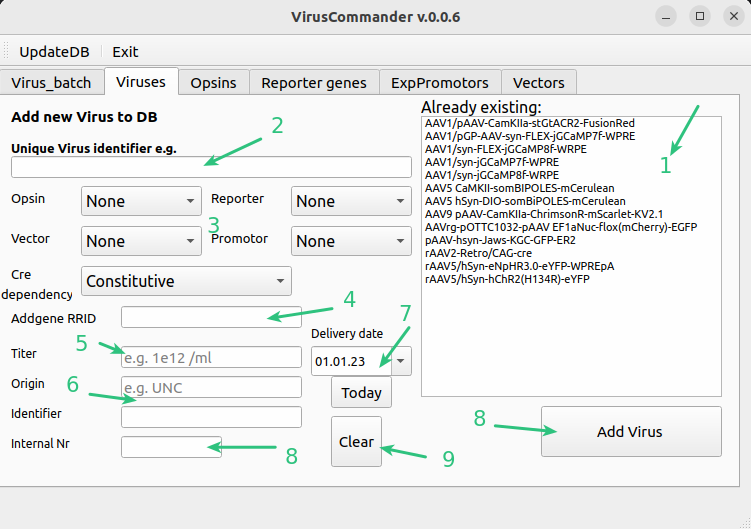
List of viruses already in DB. Please check whether yours is already available (small variations in naming are possible)
Unique full name of the virus. Try to keep the convention similar to already added viruses
Parts of the virus, Opsin if any, Reported protein, Vector , Promotor etc. Whether the expression is dependent on cre presence.
Addgene identifier (optional)
Titer in particles/ml can be entered using 1e12 notation
Virus origin and identified of supplier
Delivery date
Our internal box id where the aliquoted virus is stored.
Add virus
Clear fields
Add new batch of an existing virus#
Here we can add a newly delivered batch of an already existing virus.
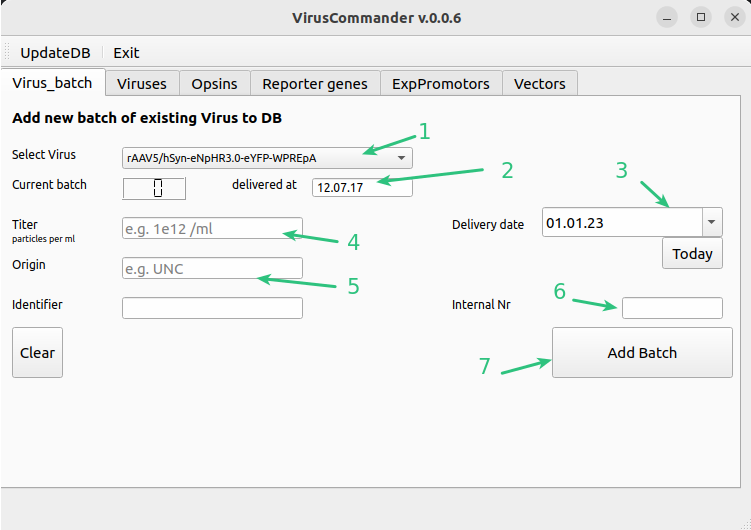
Choose a virus (has to be in DB)
Information about current batch (delivery date)
Enter new delivery date
Titer in particles/ml can be entered using 1e12 notation
Virus origin and identified of supplier
Our internal box id where the aliquoted virus is stored.
Add entry
Add opsin to DB#
Here new opsins can be entered. Here opsin is loosely defined as the functional payload of the virus and thus incldues Cre or GCaMPs.
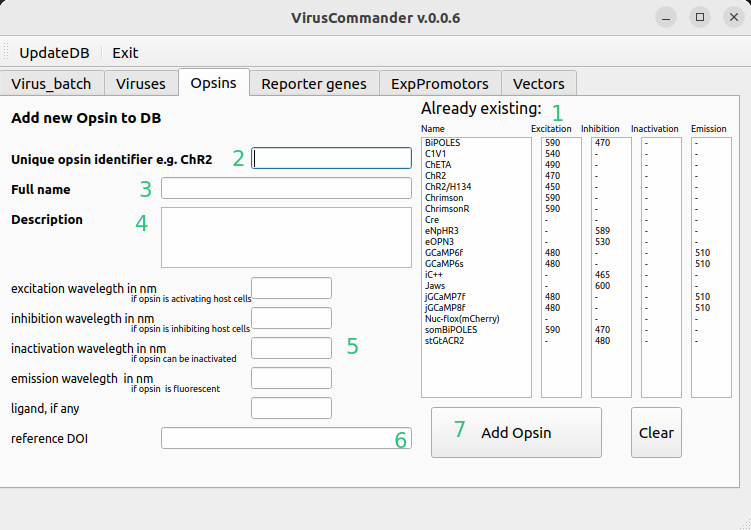
Already existing opsins
Short unique name
Full name
Description of some details
Please enter functional wavelegths for inhibition, excitation if opsins are modulating neural activity.
DOI of the paper presenting the vector (optional)
Add entry
Add new promotor#
Here new expression promotors can be added.
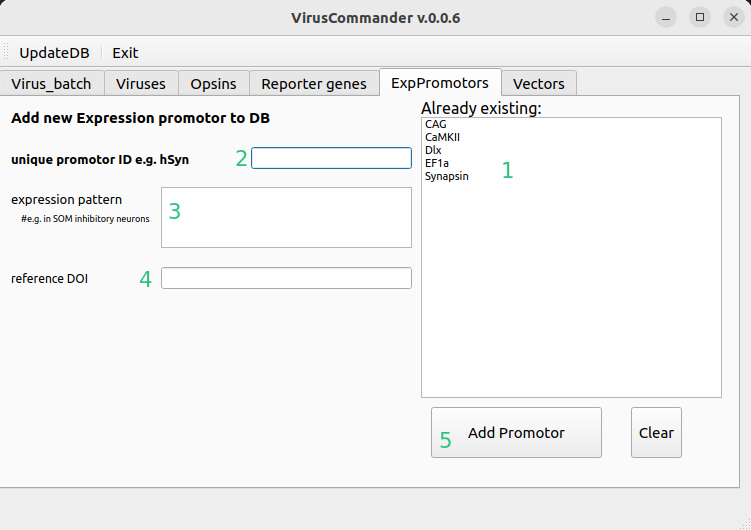
Already existing expression promotors
Short unique name
Expression pattern (optional)
DOI of the paper presenting the vector (optional)
Button to add
Add new reporter gene#
Here new reported genes (fluorescent proteins) can be entered.
Already existing reporter genes
Short unique name
Excitation wavelength
Emission wavelength (signal)
Button to add
Clear fields
Add new vector#
Here new vectors (serotypes of viruses or differen virus-types can be entered)
Already existing vectors
Short unique name
A bit more descriptive name
Some description of specific tropism of the virus if any is known. (optional)
Spread pattern of the virus
DOI of the paper presenting the vector (optional)
Button to add
Clear fields
written by: Artur
last modified: 2024-01-30